HTB 2023 – BioBundle
08 December 2023 – Written by Valentin Huber – in ctf, cyberchef, decompile, ghidra, and rev
Challenge
We’ve obtained a sample of zombie DNA. Can you extract and decrypt their genetic code - we believe we can use it to create a cure…
Solution
I first threw the binary into Ghidra. Turns out, there are two interesting functions, main
undefined8 main(void)
{
int res;
undefined8 retval;
size_t offset;
char input [128];
code *virtual_file;
undefined8 handle;
handle = get_handle();
virtual_file = (code *)dlsym(handle,&DAT_00102019);
if (virtual_file == (code *)0x0) {
retval = 0xffffffff;
}
else {
fgets(input,0x7f,stdin);
offset = strcspn(input,"\n");
input[offset] = '\0';
res = (*virtual_file)(input);
if (res == 0) {
puts("[x] Critical Failure");
}
else {
puts("[*] Untangled the bundle");
}
retval = 0;
}
return retval;
}
and get_handle.
long get_handle(void)
{
long i2;
undefined8 *puVar1;
byte bVar2;
byte decrypted;
undefined8 some_var1;
undefined8 some_var2;
undefined8 some_buff [511];
long res;
uint virtual_file;
ulong i1;
bVar2 = 0;
virtual_file = memfd_create(&DAT_00102004,0);
if (virtual_file == 0xffffffff) {
/* WARNING: Subroutine does not return */
exit(-1);
}
for (i1 = 0; i1 < 0x3e08; i1 = i1 + 1) {
decrypted = __[i1] ^ 0x37;
write(virtual_file,&decrypted,1);
}
some_var1 = 0;
some_var2 = 0;
puVar1 = some_buff;
for (i2 = 0x1fe; i2 != 0; i2 = i2 + -1) {
*puVar1 = 0;
puVar1 = puVar1 + (ulong)bVar2 * -2 + 1;
}
sprintf((char *)&some_var1,"/proc/self/fd/%d",(ulong)virtual_file);
res = dlopen(&some_var1,1);
if (res == 0) {
/* WARNING: Subroutine does not return */
exit(-1);
}
return res;
}
Let’s see what is happening, starting with the initial call to get_handle:
memfd_createcreates a virtual file in memory.- Then, in the first loop, data is loaded (from
__in the.datasection of the binary), decrypted (XORed with0x37) and stored in the virtual file. - Some other (from what I can tell irrelevant) code is executed.
- Finally, control of the virtual file is returned to the
mainfunction.
There:
- The virtual file is loaded.
- The user is asked for an input.
- The data in the virtual file is then called as a function, with the user input as the argument.
- The output of this function determines if the user input was correct.
Based on this, we can extract the data from the outer binary ourselves, decrypt it, and further analyze it. Mark the section in Ghidra, right click, Copy Special, Python List. Then, a short python script to decrypt it and save the raw binary to a file:
input = [ 0x48, 0x72, 0x7b, #...
input = [e^0x37 for e in input]
input = bytes(input)
with open("biobundle_internal", "wb" ) as f:
f.write(input)
We’ve got a new binary, so Ghidra it is once more. There is no main function this time, but I found the following:
bool _(char *param_1)
{
int iVar1;
undefined8 local_48;
undefined8 local_40;
undefined8 local_38;
undefined8 local_30;
undefined8 local_28;
undefined8 local_20;
undefined8 local_18;
undefined8 local_10;
local_48 = 0x743474737b425448;
local_40 = 0x5f3562316c5f6331;
local_38 = 0x6c3030635f747562;
local_30 = 0x7d7233;
local_28 = 0;
local_20 = 0;
local_18 = 0;
local_10 = 0;
iVar1 = strcmp(param_1,(char *)&local_48);
return iVar1 == 0;
}
A strcmp call and some hardcoded values. Cyberchef’s turn:
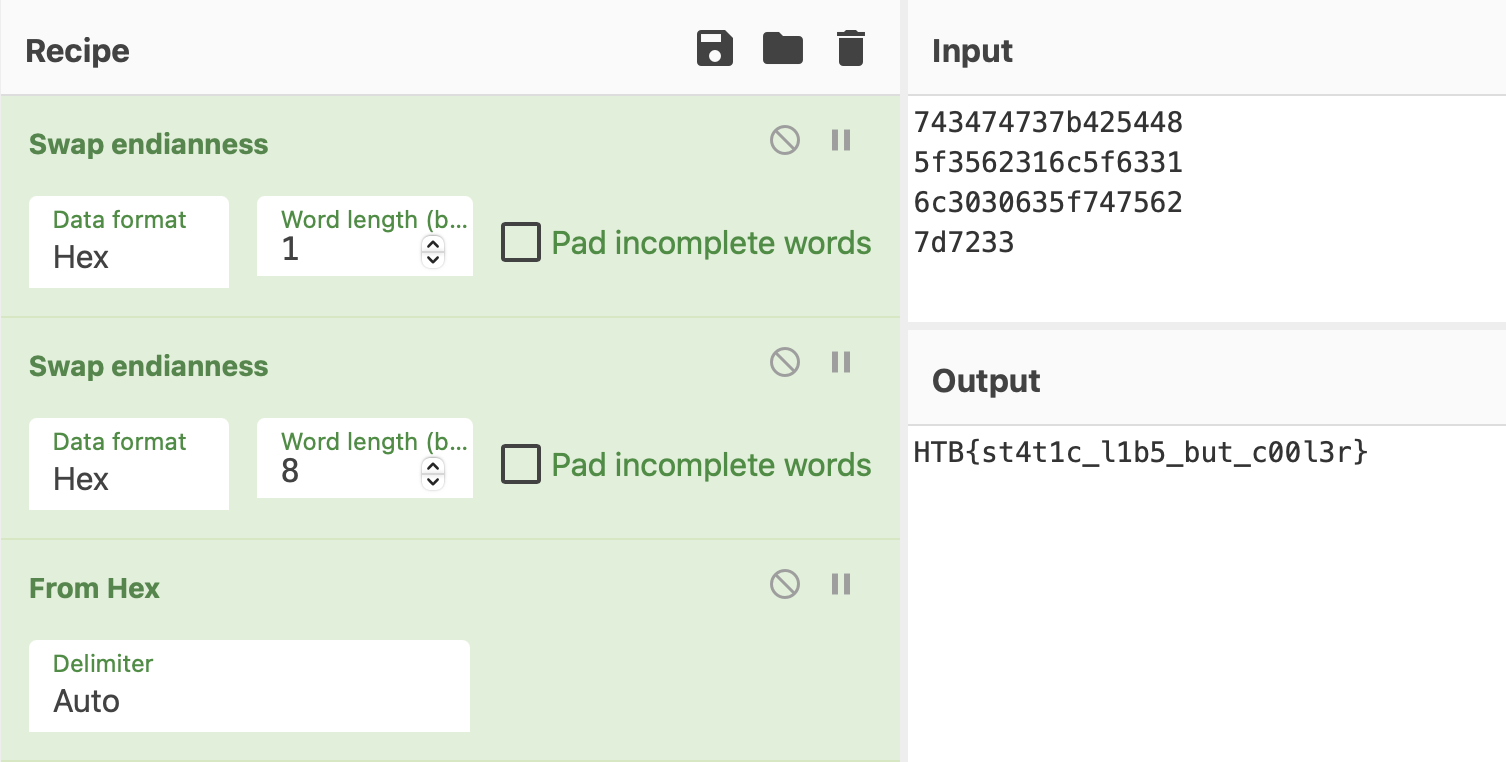
HTB{st4t1c_l1b5_but_c00l3r}